Research program : Hereditary neuromuscular diseases
Home > Team 5 > Research program > Axe 2
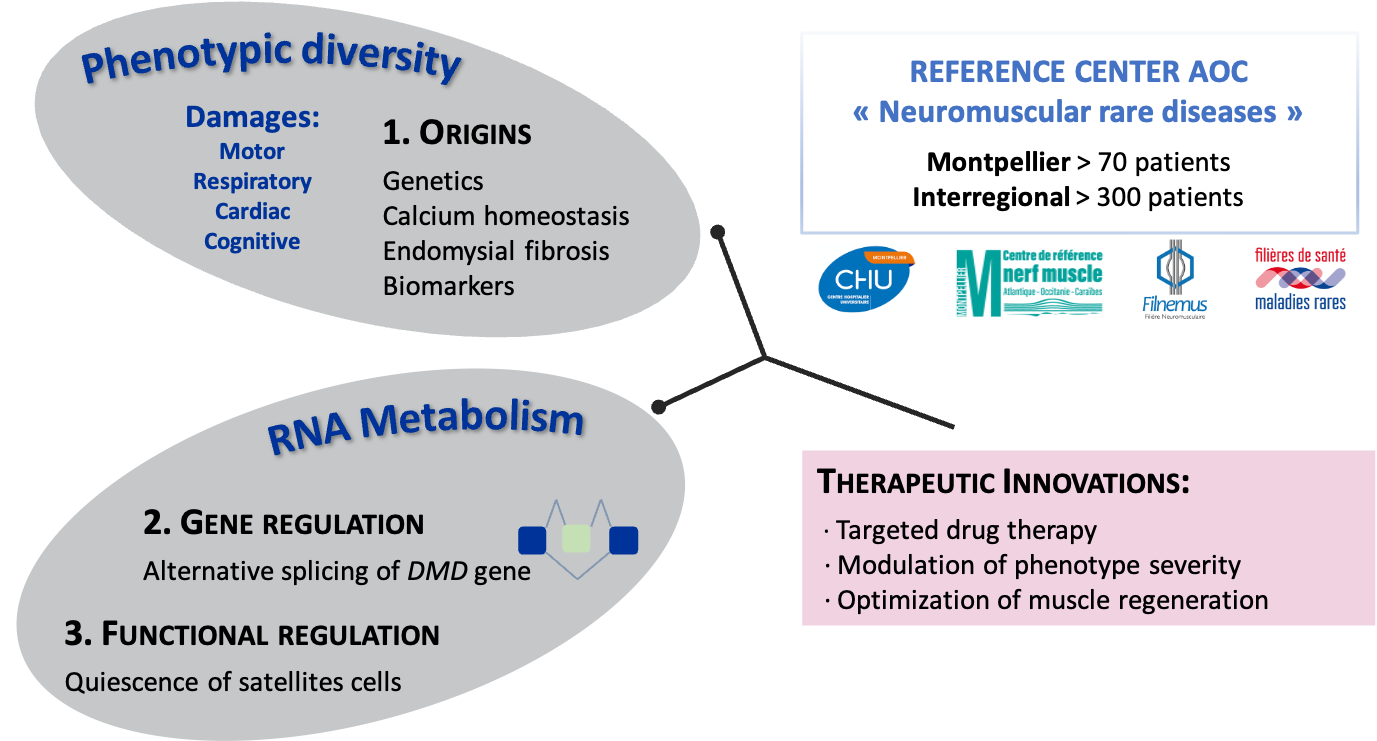
Dystrophinopathies: phenotypic diversity and gene regulation
C. Notarnicola, S. Tuffery-Giraud, J. Miro, P. Meyer, M. Cossée, F. Rivier
The dystrophinopathies are a group of different clinical forms of muscular dystrophy due to mutations in the DMD gene including Duchenne muscular dystrophy (DMD, severe form) and Becker muscular dystrophy (BMD, moderate form).
The objective of our research project is to study in the normal and/or pathological muscle cell the molecular and cellular mechanisms involved in (1) the clinical heterogeneity observed in patients, (2) the regulation of DMD gene expression, and (3) the function and activation of muscle stem cells. These approaches may ultimately contribute to the identification of biomarkers and the development of targeted therapies.
Origin of the phenotypic diversity
The clinical history of DMD, although seemingly uniform, hides a wide phenotypic variability, independent of the type of DMD gene mutations which all lead to an absence of dystrophin in skeletal muscle (Humbertclaude et al. 2012). A multicentre INSERM-DGOS study (FEDmd study) is being conducted to explore the different mechanisms (calcium homeostasis, muscle endomysial fibrosis, muscle regeneration processes) that may contribute to the observed clinical variability (Meyer et al. 2021). Our project also aims to identify relevant diagnostic and/or prognostic biomarkers for clinical follow-up or evaluation of the response to treatments in DMD, in particular circulating myomiRs, through a longitudinal study conducted in a cohort of patients (BioDystromiRs project).
Molecular mechanisms of DMD gene splicing
Alternative splicing of pre-messenger RNAs is an essential process of gene expression, which relies on the coordinated recognition of multiple signals recruiting RNA Binding Proteins (RBPs) that activate or inhibit splicing according to cellular needs. Our project aims at studying the regulation of splicing of the DMD gene (2.3 Mb, 79 exons, 7 tissue-specific promoters):
- In the physiological context: using a targeted RNA-seq approach, we have demonstrated the absence of alternative splicing in the transcript expressed in adult skeletal muscle in contrast to what is observed in other dystrophin isoforms, notably cerebral isoforms (Bougé et al. 2017). A functional screen by RNAi allowed us to identify RBPs regulating the inclusion of DMD exons in the muscle transcript (Miro et al. 2020). Our goal is now to explore the molecular mechanisms involved through an approach integrating bioinformatics, functional and biochemical analyses in order to better understand the spatio-temporal regulation of DMD gene expression.
- In the pathological context, we are interested in the role of alternative splicing as a modifier of disease severity in DMD patients (Tuffery-Giraud et al. 2017). In particular, we are investigating the underlying mechanisms of nonsense mutation-associated exon skipping that allows partial re-expression of dystrophin and attenuation of the phenotype in patients (Miro et al. 2015). Our aim is to identify RBPs (especially from the SR protein family) that can be targeted in pharmacological approaches to potentiate the spontaneous exon skipping observed in patients and improve phenotype correction.
Regulation of satellite cell function
Skeletal muscle has a remarkable capacity to regenerate after a severe injury due to the presence, under the basal lamina of the myofibrils, of muscle stem cells called satellite cells (SC). Some degenerative pathologies such as DMD affect the function of SCs and consequently impair muscle regeneration. Our research project aims to study the intrinsic mechanisms that regulate the function of adult muscle stem cells:
- Under physiological conditions, we are particularly interested in the quiescence of SCs, a stage required for the long-term survival of the stem cell pool and the identity of muscle tissue. Indeed, in adult muscle, SCs are quiescent but are also able to rapidly activate the cell cycle and the myogenic program, a function that requires specific regulation of messenger RNA. Our objective is to identify the molecular mechanisms that control mRNA translation (RBP) to maintain SC quiescence but also the signaling pathways involved in global protein synthesis.
- In the pathological context: To perform their functions, SCs must maintain a dynamic balance between different cellular states. We have shown the role of retinoic acid and associated signaling pathways on the regulation of precursor muscle cell immaturity (El Haddad M. et al. 2017). By transcriptomic analyses on the different myogenic stages, we have defined gene expression profiles from DMD patients. Our goal is now to study the function of genes that regulate SC fate in dystrophic muscle in order to develop therapeutic approaches to preserve SC function and consequently muscle regeneration.
Fundings